Jessica Brown

Associate Professor of Chemistry & Biochemistry
Contact
451 Stepan Chemistry Hall
jbrown33@nd.edu
574-631-6486
Research Cluster
Roles of RNA Triple Helices
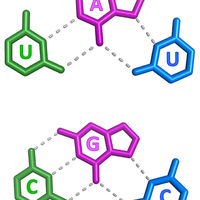
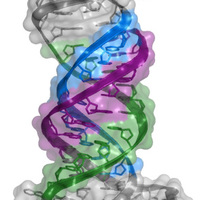
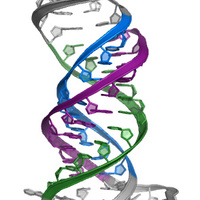
RNA structure is largely viewed as being single stranded or double stranded, although triple-stranded RNA structures were deduced to form in test tubes almost 60 years ago. Despite this early discovery, only a handful of RNA triple helices have been validated in eukaryotic cellular RNAs. The long-term goal of the Brown laboratory is to understand the biophysical, biochemical, and structural roles of RNA triple helices using the MALAT1 triple helix as a model. This triple helix forms at the 3' end of a cancer-promoting long noncoding RNA, MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), whereby a U-rich internal loop within a stem-loop structure associates with a downstream 3-terminal A-rich tract. The resulting triple helix is composed of nine U•A-U triples separated by a C+•G-C triple and C-G doublet, which altogether protects MALAT1 from an uncharacterized rapid nuclear RNA pathway.
The fundamental biophysical, biochemical, and structural properties of RNA triple helices remain to be rigorously characterized. The Brown laboratory is interested in several key questions. What is the relative stability of canonical (U•A-U and C•G-C) versus non-canonical base triples? Can successive non-canonical base triples form a stable triple helix? What are the structural parameters of an ideal RNA triple helix? What is the folding pathway of an RNA triple helix? Do proteins bind specifically to the MALAT1 triple helix? Is there an undiscovered class of triple-stranded RNA binding proteins? How does the cell degrade a highly stable triple-helical RNA structure? To investigate these questions, we are currently using a variety of approaches, mainly X-ray crystallography alongside classic biochemical and biophysical methods. Ultimately, these mechanistic studies of the MALAT1 triple helix will advance our understanding of MALAT1 and its role in cancer.
Publications
- "Molecular structure of a U•A-U-rich RNA triple helix with 11 consecutive base triples" Ruszkowska, A.; Ruszkowski, M.; Hulewicz, J.P.; Dauter, Z.; Brown, J.A. Nucleic Acids Res. 2020, 48(6), 3304-3314. DOI: 10.1093/nar/gkz1222
- "Stability of an RNA•DNA-DNA triple helix depends on base triplet composition and length of the RNA third strand" Kunkler, C.N.; Hulewicz, J.P.; Hickman, S.C.; Wang, M.C.; McCown, P.J.; Brown, J.A. Nucleic Acids Res. 2019, 47(14), 7213-7222. DOI:10.1093/nar/gkz573.
- "Structural insights into the RNA methyltransferase domain of METTL16" Ruszkowska, A.; Ruszkowski, M.; Dauter, Z.; Brown, J.A. Sci. Rep. 2018, 8(1), 5311. DOI: 10.1038/s41598-018-23608-8
- "Methyltransferase-like protein 16 binds the 3'-terminal triple helix of MALAT1 long noncoding RNA" Brown, J.A.; Kinzig, C.G.; DeGregorio, S.J.; Steitz, J.A. Proc. Natl. Acad. Sci. USA 2016, 113(49), 14013-14018. DOI: 10.1073/pnas.1614759113